Automated integration of structural, biological and metabolic similarities to improve read-across
Main Article Content
Abstract
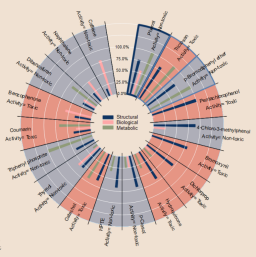
Read-across (RAX) is a popular data gap-filling technique that uses category and analogue approaches to predict toxicological endpoints for a target. Despite its increasing relevance, RAX relies on human expert judgement and lacks a reproducible and automated protocol. It also relies only on structural similarity for identifying the analogues, while other aspects are often neglected. In this paper, we propose an automated procedure for the selection of analogues for data gap-filling. Analogues were identified by a decision algorithm that integrates three similarity metrics, each considering different toxicologically relevant aspects (i.e., structural, biological and metabolic similarity). Structural filters based on the presence of maximum common substructures (MCS) and common functional groups (FGs) were applied to narrow the chemical space for the analogue search. The procedure has been implemented as a workflow in KNIME and is freely available. The workflow provides informative tabular and graphical outputs to support toxicologists and risk assessors in drawing conclusions based on the RAX approach. The procedure has been validated for its predictive power on two datasets related to high-tier in vivo toxicological endpoints, i.e., human hepatotoxicity and druginduced liver injury (DILI). The validation results gave good accuracy values (i.e., up to 0.79 for the binary hepatotoxicity classification and up to 0.67 for the three-class DILI classification) that were higher than those returned by RAX based on the sole use of structural similarity. Results confirmed the suitability of the procedure as a source of data to support regulatory decision-making.
Article Details

This work is licensed under a Creative Commons Attribution 4.0 International License.
Articles are distributed under the terms of the Creative Commons Attribution 4.0 International license (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium, provided the original work is appropriately cited (CC-BY). Copyright on any article in ALTEX is retained by the author(s).